Scientists from the University of Oxford have developed a DNA sequencing technique to help cancer cell research specialists understand tumour growth
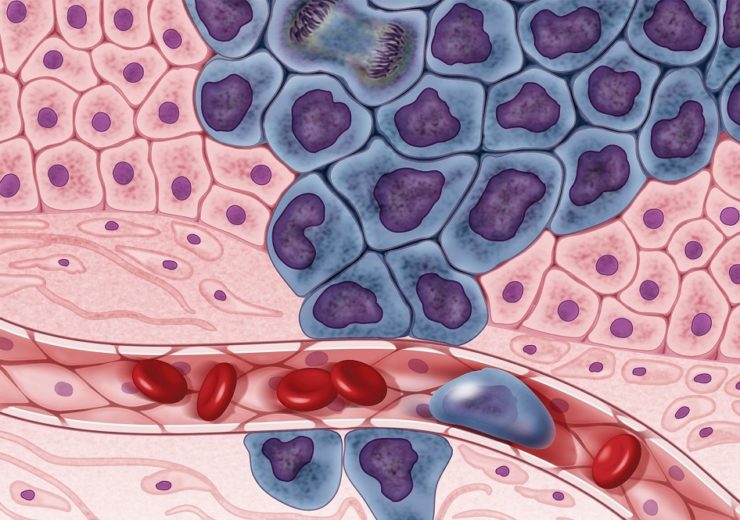
Illustration of growing cancer cells (in purple) are surrounded by healthy cells (in pink), illustrating a primary tumor spreading to other parts of the body through the circulatory system. (Credit: Darryl Leja/ National Human Genome Research Institute)
A new DNA sequencing technique developed by scientists from the University of Oxford will provide a “high definition” picture in cancer cell research.
A team of researchers from the Medical Research Council’s (MRC) Weatherall Institute of Molecular Medicine at the university developed the technique, which tracks genetic errors in individual cancer cells.
The process, referred to as Target-seq, will allow doctors to identify individual cells that are likely to mutate and lead to cancer tumour growth – giving them more reliable and accurate measurements.
The problem with previous cancer cell research techniques
Previous attempts to analyse individual cancer cells were inaccurate and often struggled to track more than one cell at a time.
The latest research marks the first time these DNA errors or mutations – which lead to cancer – can be analysed and tracked in thousands of individual cancer cells at the same time.
It is hoped this will make it easier for clinicians to personalise cancer treatments for patients and help to prevent the cancer returning.
Alba Rodriguez-Meira, one of the MRC researchers who worked on the project, said: “Having this information is really important because it not only gives us unique information about how tumours change over time, but also how they might respond in future to different treatments and drugs.
“No two patients will have exactly the same mixture of cancer cells with exactly the same pattern of evolution, and our technique will allow clinicians to monitor patient progression during clinical trials and ultimately customise the treatment they offer to the unique mixture of cancer cells that every cancer patient has.”

What does Target-seq mean for cancer cell research?
The Target-seq technique has so far been trialled on 15 patients suffering from leukemia and marks the first time that cancer cell researchers have directly tested individual cells.
They found that cells from the same patient and from the same cancer do not contain the same genetic errors and can accumulate a variety of mutations.
Dr Mariana Delfino-Machin, programme manager for cancer at the MRC, said: “Cancer is often thought of as a single disease, but it is actually a collective term for the abnormal growth and spread of cells, which can differ even within the same patient.
“If we are to treat cancer successfully, we need to understand exactly how all these different abnormalities behave and respond to treatment, and this is hugely challenging.
“This new technique takes us one step closer to achieving this goal.
“If we can understand the background and behaviour of individual cancer cells in a tumour, we have the potential to tailor more effective treatments to the individual in the future.”
Another Oxford-based company, Oxford Nanopore, last year made a breakthrough with its handheld DNA sequencing device, which is capable of analysing the biological structure of any object.
Its makers claim that DNA sequencing has the potential to bring “biological breakthroughs” within reach.